Research
Projects
The laboratory is interested in the relationships between structure and function of biologically important macromolecules. The lab utilizes the disciplines of X-ray crystallography and Cryo-EM to determine the atomic resolution structures of the macromolecules and higher-ordered complexes. The molecule’s atomic resolution structure provides tremendous insight into understanding how the macromolecule (protein) functions. Hypotheses on structure – function relationships can be drawn from the structures, which are investigated using biochemistry, cell biology, and other biophysical techniques. Currently, the laboratory is focusing on three important biochemical pathways; RNA editing by ADARs, sulfate activation, and sialic acid.
ADAR:
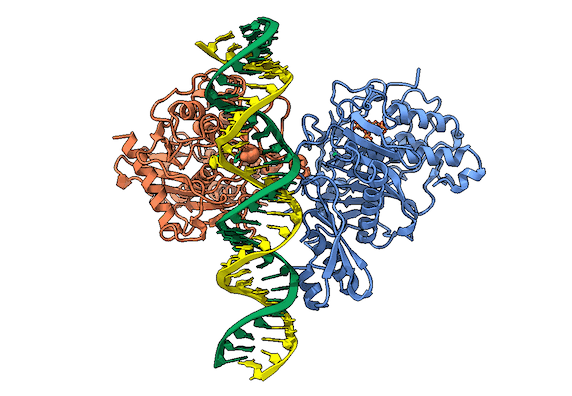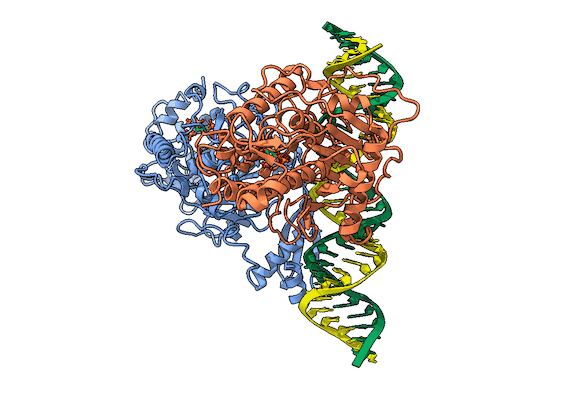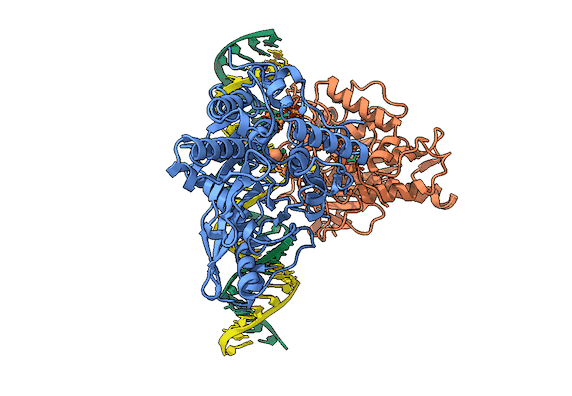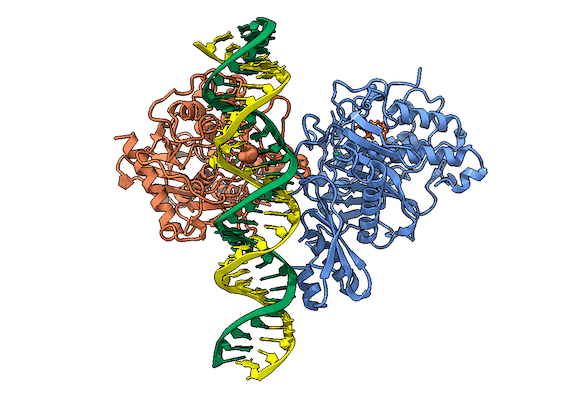
RNA is frequently edited after being transcribed by RNA polymerase. One of the most abundant RNA modifications found in metazoans is the deamination of adenosine to inosine (A-to-I). This reaction is catalyzed by the class of enzymes called adenosine deaminases acting on dsRNA (ADAR) .
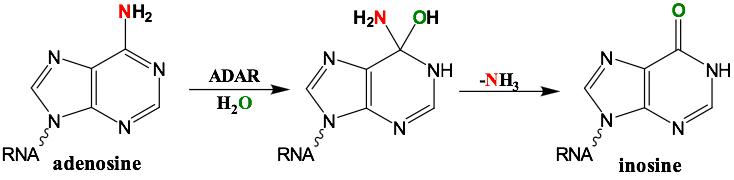
In the ADAR deamination reaction, the resulting inosine nucleotide product behaves more like guanosine in hydrogen-bonding patterns, consequently A-to-I edits can significantly alter the RNA function including; recoding mRNA, altering splice sites, and changing RNA secondary structure. Over three million RNA A-to-I edits have been documented in healthy human tissues. RNA editing is critical for normal cellular function and dysregulated ADAR activity can resulted in a variety of neurological disorders such as epilepsy and Prader Willi Syndrome, depression, schizophrenia, amyotrophic lateral sclerosis, as well as cancer
In collaboration with the Beal lab, we have structurally and biochemically characterized human ADAR2 deaminase domain bound to a 23mer RNA duplex, which contained the nucleoside analog 8-azanebularine in place of adenine at the edited site. Upon bonding in the ADAR active site, 8-azanebularine becomes hydrated mimicking the proposed intermediate thus forming a tight-binding dsRNA-ADAR complex amenable for structural studies.
We are currently continuing our studies on investigating the structural basis for RNA selectivity and mechanism by looking at different ADARs and ADAR constructs bound to RNA.
SULFATE ACTIVATION:
A project of interest in the lab is understanding sulfur assimilation, or fixation. This project focuses on the structural features underlying the catalytic and regulatory properties of the sulfate activating enzymes, ATP sulfurylase, APS kinase, and PAPS Reductase, which catalyze in order, the reactions:

where: APS = adenosine 5′-phosphosulfate (adenylylsulfate); PAPS = 3′-phosphoadenosine-5′-phosphosulfate (3′-phosphoadenylylsulfate); PAP = adenosine 3′,5′-bisphosphate, and TRX is Thioredoxin.
The first two enzymes in the pathway (ATP Sulfurylase & APS Kinase) are conserved in all organisms to activate sulfate into the universal sulfate donor (PAPS) used to sulfurylate many diverse biomolecules including proteins, lipids, and carbohydrates. The last step in sulfur assimilation, PAPS reductase, is not conserved in many higher eukaryotes, such as humans. Our lab is exploring the structural basis for sulfate activation and sulfur assimilation in many organisms including bacterial, archaea, fungi, and humans, to better understand how each organism as adapted the enzymes to suit the physiological function of the pathway. Some chemolithotrophic organisms use the pathway in the final oxidation of sulfur to harness energy. Additionally, we are investigating these enzymes in Mycobacterium tuberculosis, because many of the enzymes in the pathway display sequence and structural features different than the orthologous human enzymes, making them potential therapeutic targets to treat tuberculosis.
Sialic Acid/Glycobiology:
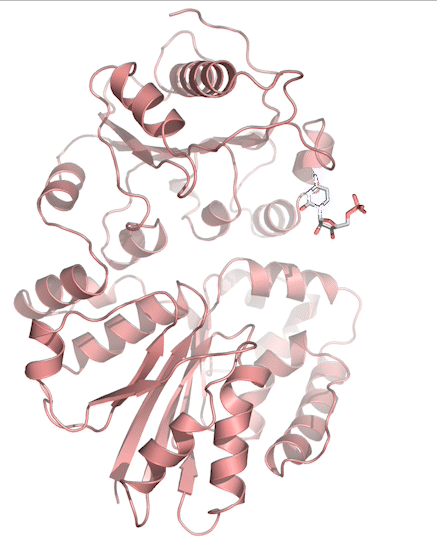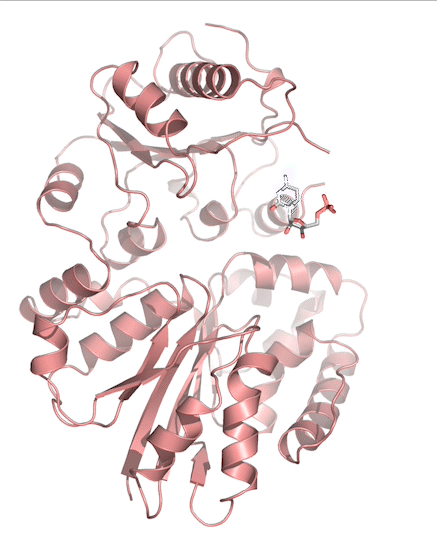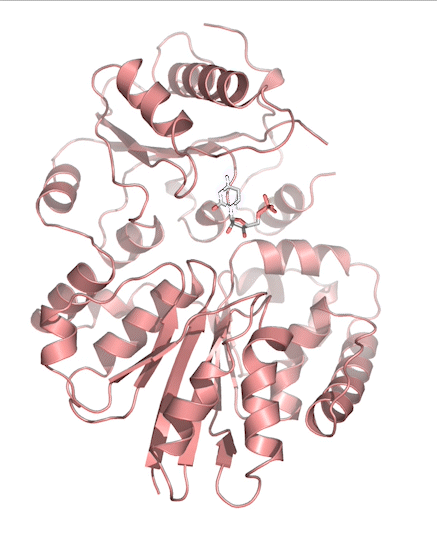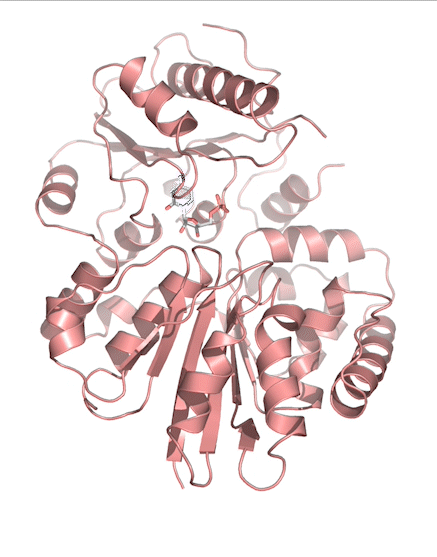
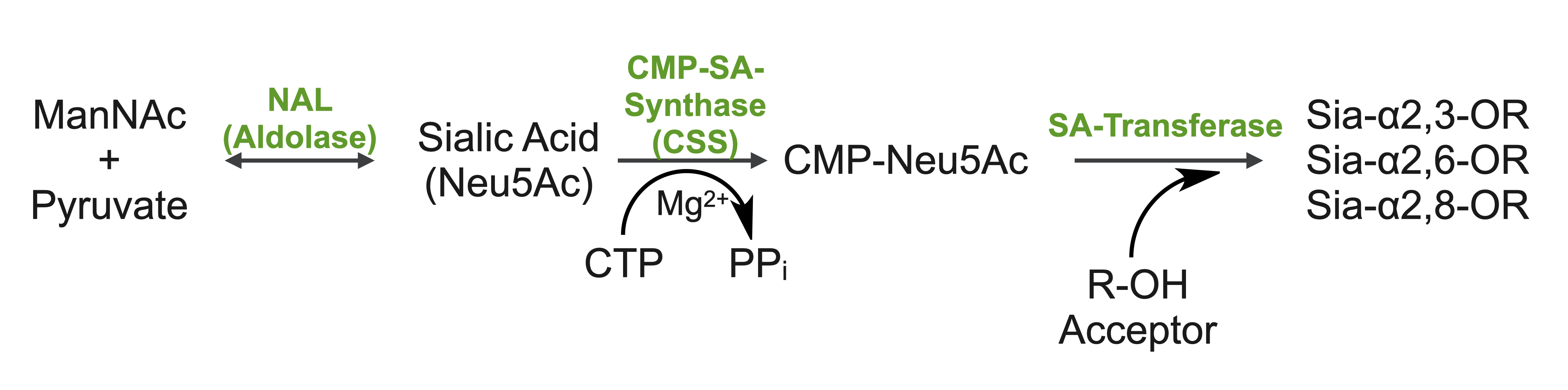
Sialic acids (N-acetylneuraminic acid) are nine-carbon acidic monosaccharides that are typically found at the terminus of cell-surface glycans. As the terminal carbohydrate residue, sialic acid is one of the first molecules encountered in cellular interactions and has been found to play important roles in cellular recognition, communication and pathogen attachment, such as influenza virus. Sialic acid has also been found on the surface of many pathogenic bacteria where they may function as molecular mimicry to evade the host’s immune system.
Our lab, in collaboration with the Chen lab, have been investigating the structure-function relationships of enzymes involved in the synthesis and conjugation of sialic acid. Our lab recently determined the crystal structures of the enzymes N-Acetylneuraminate lyase (NAL), also known as aldolase, CMP-sialic acid synthetase (CSS), and sialyltransferases, all with various substrate, product or substrate analogues.Despite their important biological functions, little is known about the molecular architecture and mechanism of these enzymes. Our goal of this project is to elucidate the molecular structure and catalytic mechanism of these enzymes with a long-term goal to find modulators to control diseases associated with both host and pathogen enzymes. Another aim is to use structure-guided protein engineering to modify the enzymes, which can be used in the chemoenzymatic synthesis of unnatural or modified sialic acid analogues for many diverse biotechnology applications.